Artemis - A “Focused Research Organization” to Establish New Mammalian Models for Predictive Preclinical Research
The next breakthroughs in complex disease will come from new animal species, not the lab mouse.
Problem Statement
For over a hundred years, we have relied almost exclusively on the lab mouse, developing nearly every tool along the way. Now, despite representing over 95% of research animals, the canonical lab mouse struggles to model complex disease of aging, visual neuroscience, and female reproductive biology. The mouse lacks key human traits, making it difficult to translate new discoveries into therapeutics. While AI and ex vivo/organoid models are becoming increasingly common for complex disease drug discovery, their predictive validity and clinical translatability remain poorly understood. Alternative mammals with closer human physiology have been characterized, but their use in research is bottlenecked by convenience, cost, and tradition. A robust experimental toolkit for non-model mammals is a public good that, if developed, would enable research in translational mammals to unlock biological discoveries of high predictive validity. Now, as we continue to face many complex diseases, we must embrace new mammals to solve them.
Project Concept
Artemis will develop the foundational toolkit for three new non-model mammals—the spiny mouse, naked mole rat, and tree shrew—to bridge the translational gap left by canonical lab rodents by enabling new systems to study complex diseases. Each of these mammals has evolved unique traits that make them exceptional for complex disease research: the only menstruating rodent, the spiny mouse (Acomys dimidiatus); the extraordinarily long-lived and cancer-resistant naked mole rat (Heterocephalus glaber); and the diurnal tree shrew (Tupaia spp.); all are critically underdeveloped but increasingly important in studying diseases of female reproduction, healthy aging, and visual neuroscience, respectively. The suite of tools we will develop, including species-specific antibodies, fully annotated genomes, transgenics, and a community web portal, will be distributed to the public through independent deployment and industry partnerships. Open access, community engagement, and resource-sharing across academic and industry are also key outcomes of Artemis, made possible only through a FRO model. These tools will provide new methods to understand the underlying physiology of more than three diverse complex disease areas and unlock novel targets for therapeutics. By enabling the use of these animals, Artemis aims to spark a broader shift in how scientists study these diseases.
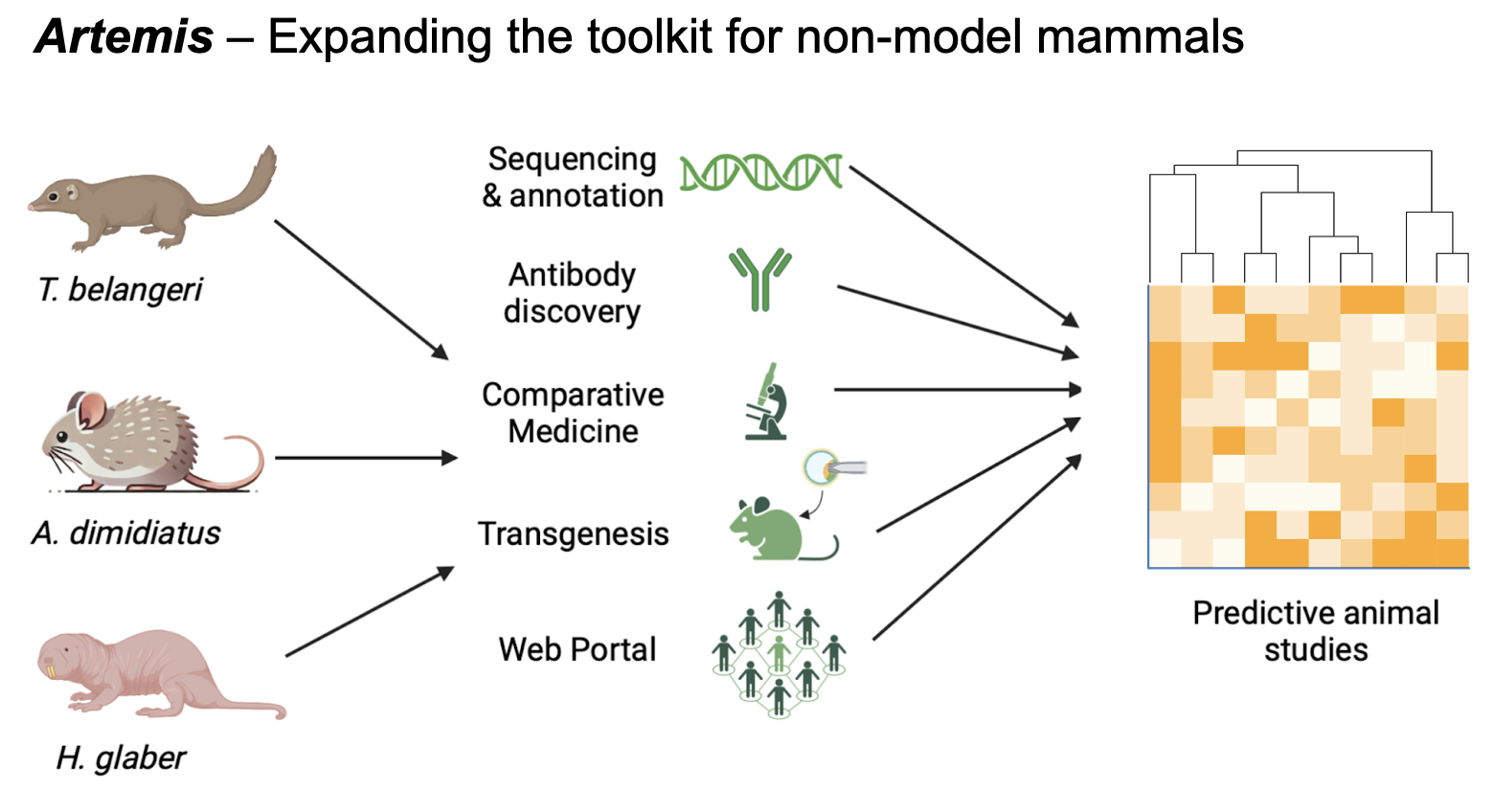
Project Deliverables
- Antibodies – Species-specific antibodies are a critical resource that is lacking across non-model organisms. Academic scientists currently face the time-consuming task of individually identifying effective antibodies by screening those from mice, humans, and adjacent species. Many animal communities like the Acomys also have private sheets that circulates through trusted academics listing effective antibodies. However, non-specific antibodies often cannot detect unique molecules of different species, which significantly reduces accessibility as a result. We therefore propose the parallel repurposing and de novo synthesis of tissue-specific antibodies across all three animal programs, with an initial emphasis on tissue-types with the highest translational potential such as the Acomys endometrium, Heterocephalus fibroblasts, and Tupaia retina. These standard antibodies will be developed and licensed in collaboration with partners such as Thermo Fisher, with protocols being continuously uploaded on our portal for all scientists to access and comment on.
- Full genome sequencing and annotation – Genome assembly and annotation of functional sequences is a critical milestone that will enable novel research in the species through functional genomic experiments in both academia and industry. While non-model organism sequencing efforts are ongoing or completed, it is the annotation of genes and regulatory elements that makes this information truly useful. We will leverage advanced bioinformatics tools and experienced scientists to accurately identify and annotate genes, pathways, and functional elements. We plan to make this information freely available through resources like Ensembl and GenBank.
- Transgenic (Cre-Dependent Cas9 Knock-in) animal – Transgenic animals are a critical resource for genetic overexpression and knock-down for novel biological pathway discovery. Genetic models remain the preferred method of challenge to induce disease models in industry, and developing these systems will greatly facilitate the direct translational impact of the respective species for drug development. We therefore propose the development of Cre-dependent Cas9 knock-in animals to enable precise and controlled genetic modifications. Such a model will be instrumental in elucidating complex gene functions and interactions, further improving the species’ utility in understanding complex diseases.
- Comparative medicine – Biological breakthroughs from animals have historically been products of serendipity. The discovery of insulin through the beagle, fluorescent proteins from jellyfish, nanobodies from camelids, ACE inhibitors from snakes, all occurred from serendipitous and long-term research on fundamental biomolecular properties of unique animals. While developing standard housing and care protocols for the broader research community, we aim to systematically characterize and publicize the biomolecular properties of core animal phenotypes under our care, inspiring novel applications of these models.
- Web Portal – A centralized portal will serve as the intellectual resource hub to facilitate open collaboration and discussion, serving a major unmet need for the scattered non-model mammal community. This portal will house much of Artemis’ key deliverables including comprehensive protocols, methods, and more. By consolidating these resources, the portal aims to foster a vibrant, collaborative community, accelerate scientific discoveries, and ultimately advance the use of non-model mammals in research around the world.
Appendix
What is a Focused Research Organization?
Focused Research Organizations (FROs) are mission-driven, time-bound research teams structured similarly to startups. Designed to address specific, medium-scale scientific or technological problems, FROs develop breakthrough tools, technologies, processes, or datasets that function as public goods, expanding the capabilities of the broader research ecosystem and accelerating overall progress. These projects often address needs that are overlooked by traditional funding systems due to misaligned incentives, bureaucratic hurdles, or differing institutional priorities.
Why does this project need to be a FRO? Why is it not a good fit for traditional research organizations (academic labs, startups, etc.)?
High-throughput tool development and distribution is unprecedented in academia due to limited funding, talent, incentives, and the long timescales required. Conversely, private companies are not incentivized due to limited investment returns, lack of precedent for ever-conservative pharma, and logistical challenges around new animal onboarding. For tools like antibodies to be distributed effectively, we cannot discover them individually and license them out to large vendors. Rather, establishing partnerships with existing vendors and their antibody pipeline would be the most effective way to distribute the tools to scientists. These partnerships are rarely done in academia or startups since the technical requirements and financial incentives often do not align. Recent examples of non-profit, coordinated organizations include Cultivarium and YCharos. This project also involves extensive community engagement in each non-model animal community but also the research communities adjacent to them. Establishing new animals in a field takes time, and scientists are inherently skeptical. Engaging all the communities while developing tools by attending conferences, hosting seminars, and distributing information through the web portal is a consortium-like approach that is best done through the FRO style.
How This Project Will Benefit Scientific Progress
Many complex diseases of aging, neuroscience, and female reproduction, are difficult to model in canonical lab rodents, limiting the discovery of novel disease-modifying therapies. By developing robust toolkits for high-potential species with human-like traits, Artemis will enable more predictive, reproducible studies across academia and industry. Today, the largest bottlenecks preventing the use of better animal models are not scientific interest, but infrastructure: the lack of antibodies, genetic tools, annotated genomes, and standardized protocols. These foundational resources are what allow a model to be adopted across labs. We directly addresses this gap by systematically developing and distributing essential tools for three underleveraged mammalian models. These resources will be validated and made available through open-access platforms and partnerships, lowering the barrier to entry for researchers and enabling comparative studies that were previously impractical or impossible.
By making these new models accessible and usable at scale, we will empower a broader scientific community to be able to ask more questions, develop more robust disease models, and ultimately generate more translatable discoveries.
Frequently Asked Questions
How and why did you select these animals? Why not select the best models with a less-biased, high-throughput screen?
The selection process began with developing a comprehensive shortlist, generated through informal surveys and direct polling of scientists at multiple research institutes. Researchers were invited to nominate non-traditional mammalian models they believed had significant promise for translational research. This shortlist was then evaluated using a weighted, multi-criteria analysis. Key factors included:
• Robustness of the research community: Consideration of whether an active and collaborative network of researchers already exists or can be rapidly mobilized around a given model.
• Publication history: Review of the number and quality of peer-reviewed publications involving each candidate species, indicating both the maturity and depth of prior research.
• Projected translational impact: Assessment of the species’ physiological or genetic similarities to humans and their relevance to disease mechanisms that are poorly modeled in standard laboratory animals (Mus, Rattus, etc).
• Shared resource gaps: Identification of existing bottlenecks, such as the lack of standardized reagents, protocols, or breeding colonies, where focused investment could rapidly accelerate research progress.
This approach allowed us to select a cohort of animal species that balanced maximum potential translational impact with ease of adoption by the broader scientific community.